Integrations of endogenous retroviruses have previously been detected in mouse and human genomes. While most are inert, a subset can affect the activity of host genes. Using wild-type mouse embryonic stem cells derived from different genetic backgrounds, Prof. Danny Leung and his research team identified thousands of potential polymorphic ERVs. In particular, they have discovered a polymorphic element MMERGLN capable of changing the activities of neighboring genes. Their work published in Nature Communications has shed light on how the relics of viruses can shape our epigenome and give rise to different phenotypes between individuals.
Endogenous retroviruses (ERVs) are repetitive elements that make up approximately 8% and 10% of the human and mouse genomes, respectively. While some have remained intact throughout evolution, most of the ERVs in mammalian genomes have been reduced to fragments of the original. These elements can disrupt proper transcriptional regulation and cause genome instability in the host. Therefore, defense mechanisms, including epigenetic pathways, work to suppress ERVs and reduce their harmful potential.
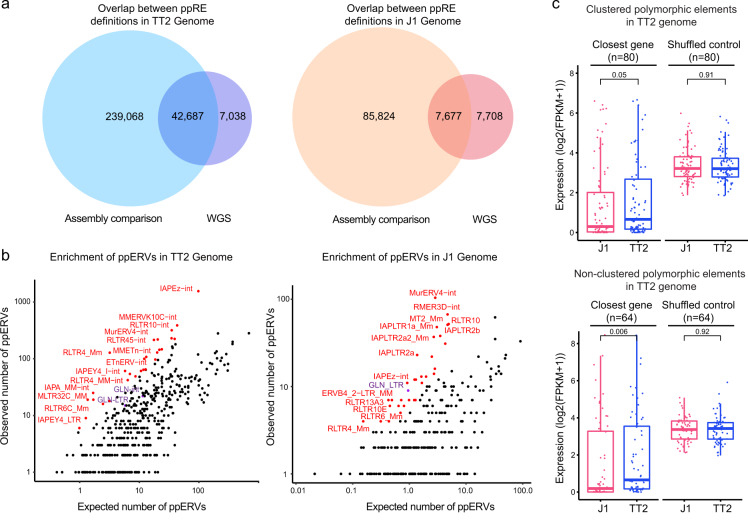
Interestingly, a subset of ERVs has escaped silencing by participating in normal molecular functions within the host. For example, they can contribute to new protein-coding sequences or regulate gene expression in different cell types. To find out the effects of ERVs, the research team analyzed murine embryonic stem cells (mESCs) from two commonly used laboratory mouse strains: J1, derived from the 129S4/SvJae strain and TT2, derived from C57BL/6×CBA/J F1 embryos.
The Leung lab discovered a full-length MMERGLN (abbreviated as GLN) provirus that integrates near the Klhdc4 gene in strains C57BL/6 and CBA/J but is absent in other genetic backgrounds. Using epigenetic tools, they revealed that this polymorphic GLN contributes to the strain-specific transcriptomic properties, including the ATF4 transcription factor associated with differential stress responses among mouse strains. Their results have demonstrated how naturally occurring polymorphic ERVs can regulate strain-specific transcriptional networks and chromatin states. Using genome-wide analysis, the research team has also found that this phenomenon is much more common than previously thought.
Journal Reference:
Zhou X, Sam TW, Lee AY, Leung D. Mouse strain-specific polymorphic provirus functions as cis-regulatory element leading to epigenomic and transcriptomic variations. Nat Commun. 2021 Nov 9;12(1):6462. doi: 10.1038/s41467-021-26630-z.10.1038/s41467-021-26630-z.