Professor Li-Sheng ZHANG, (Division of Life Science and Department of Chemistry, HKUST) and his research team have recently made a significant breakthrough in cancer detection by introducing a new RNA modification method called LIME-seq (Low-Input Multiple Methylation Sequencing) which has been published in Nature Biotechnology. This innovative approach focuses on analyzing circulating free RNA (cfRNA) in the blood, which has the potential to improve early cancer diagnosis, particularly for challenging cases like colorectal cancer.
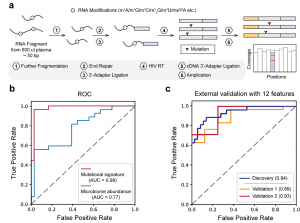
Overview of the LIME-seq workflow. b, Receiver operating characteristic (ROC) curves support the superior performance of mutation signatures compared to microbiome abundance obtained from LIME-seq. c, ROC curves showing that 12 microbiome-derived biomarkers on cfRNA distinguish patients with CRC from noncancer-bearing controls across discovery (that is, training) and two validation cohorts.
Liquid biopsy technology has become a key method for diagnosing and monitoring cancer. It involves testing for biomarkers in the blood, such as circulating free DNA (cfDNA) and cfRNA. While cfDNA testing has gained traction, it often struggles to detect early-stage cancers like colorectal cancer (CRC) due to the low levels of cfDNA present. This has prompted scientists to explore cfRNA as a promising alternative.
cfRNA consists of tiny RNA fragments found in the blood, which are released from dying cells or tissue turnover. Although cfRNA has historically been difficult to work with due to its fragility and low quantity, it offers unique advantages. It provides insights into gene activity and can signal early changes related to cancer, making it potentially more useful than traditional DNA tests.
LIME-seq stands out for its ability to detect RNA modifications with minimal sample input—requiring less than 2 ng of RNA. The researchers demonstrated that this method could identify various RNA changes linked to cancer. In their studies, they analyzed plasma samples from colorectal cancer patients and found elevated levels of microbial-derived cfRNA methylation, leading to the development of a highly accurate model for distinguishing cancer patients from healthy individuals.
LIME-seq has three major highlights:
The findings from this research highlight how cfRNA modifications could be used as non-invasive tools for early cancer detection. The connection between microbial cfRNA and tumor status opens new possibilities for understanding how the human microbiome interacts with cancer. This innovative approach not only promises advancements in cancer diagnostics but also paves the way for exploring other diseases, making it a significant step forward in medical research and patient care.
Publication reference
Ju, CW., Lyu, R., Li, H. et al. Modifications of microbiome-derived cell-free RNA in plasma discriminates colorectal cancer samples. Nat Biotechnol (2025). https://doi.org/10.1038/s41587-025-02731-8