All living creatures, from simple unicellular yeast to complicated human beings, propagate themselves through cell divisions. Each division requires the exact replication of the genomic DNA, which are the blueprints and identities of every organism. During DNA replication, the origin recognition complex (ORC), a DNA binding complex with six subunits, binds to the origins of replication, where DNA replication is initiated.
In eukaryotes (any cell or organism that possesses a clearly defined nucleus), the location and timing of DNA replication are tightly-controlled under normal conditions. However, the mechanisms that govern DNA replication are not universal across all eukaryotes, and the question of how such differences have come about has fascinated scientists for decades.
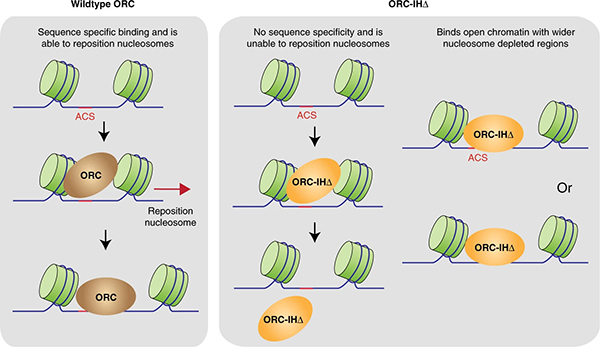
ORC is highly conserved in protein structure and function from yeast to human, but the DNA sequences of the replication origins recognized by human ORC exhibit no specific common features, which is drastically different from its yeast counterpart. Prof. Bik-Kwoon Tye and Prof. Danny Leung, in collaboration with the University of Hong Kong (HKU), recently made a breakthrough on how the ORC selects the site for origins of replications and the basis behind the divergence between humans and yeast.
The research team discovered that the selectivity determinant of ORC for DNA binding lies in a 19-amino acid insertion helix in the Orc4 subunit (Orc4-IH), which is present in yeast but absent in humans. By removing this Orc4-IH motif, the yeast ORC becomes “humanized” and selects origins based on chromatin landscape (genomic nucleosome profile) instead of base-specific binding at defined locations.
“Structure informs function,” said Prof. Tye. “In 2018, we solved a high-resolution structure of the yeast ORC using cryo-electron microscopy. When we compared the structures between yeast and human, we found one important difference between them, which lies in one subunit of ORC-Orc4.”
“The yeast Orc4 has additional 19-amino acid which makes contacts with origin DNA and is not present in human. We immediately realized that this short fragment of Orc4 might be a critical factor that causes the different behaviors of ORC in DNA binding between yeast and human. We then decided to delete this fragment in yeast cells to test if it could convert the yeast ORC into one with human properties. Indeed, the engineered ORC in yeast behaves more like a human ORC, or as we call it, a humanized ORC.”
DNA replication is one of the most fundamental processes in cells. In yeast cells, DNA replication starts from a set of sites, termed replication origins. These sites share similar features with specific DNA sequences recognized by the ORC subunit complex formed by Orc1-6. ORC binds to origin DNA, then serving as a platform to recruit Mcm2-7 helicase to open up double-stranded DNA and provide templates for DNA replication.
“The activities of each origin are tightly regulated to ensure genome integrity,” said Prof. Yuanliang Zhai, the collaborator of the study and Assistant Professor at the School of Biological Sciences, Faculty of Science at HKU. “Mis-regulation of replication initiation can cause either under- or over-replication of chromosomal DNA, which will induce DNA strand breaks, gross chromosomal rearrangements, and genome instability, a characteristic of almost all human cancers. Our studies lay a solid foundation to identify pairs of interactions, that are critical for origin recognition and helicase loading, with the potential as targets for anticancer drug screening and design.”
Inhibition of replication initiation is considered as an effective anti-cancer strategy for the selective killing of cancer cells through apoptosis while normal human cells arrest in G1 state (growth) or withdraw from the cell cycle into G0 state (inactive).
“This study demonstrates the power of multi-disciplinary approaches to answer fundamental questions in life sciences.” said Prof. Leung. “It began with the atomic model of the yeast ORC bound to origin DNA and the discovery that a single motif in one of the subunits is responsible for the base-specific recognition of DNA by ORC. We conducted genome-wide assays and biochemical experiments to define binding characteristics, which led to the model that removal of this motif is the basis for the behavior of the human ORC, culminating in an insight for the evolution of ORC as eukaryotes adopt more complex genomic and epigenomic structures. This insight also bears critical information on disease transformation that is often associated with the plasticity of DNA replication.”
Prof. Leung’s team was responsible for the epigenomics and bioinformatic analyses of this study. Prof. Leung is the Director of HKUST’s Center for Epigenomics Research, which coordinates the efforts of the Hong Kong Epigenomics Project and facilitates regional researchers in carrying out epigenomic studies.
Further understanding of the preferred DNA shapes and nucleosome positioning requirements will provide new insights for the plasticity of the human ORC in selecting replication initiation sites during programmed development and disease transformation. More research on this topic will also help identify potential targets for anti-cancer drug screening and therapy design.
Journal Reference:
Lee CSK, Cheung MF, Li J, Zhao Y, Lam WH, Ho V, Rohs R, Zhai Y, Leung D, Tye BK. Humanizing the yeast origin recognition complex. Nat Commun. 2021 Jan 4;12(1):33. doi: 10.1038/s41467-020-20277-y.
Source: HKUST News Release 28-Jan-2020