Skeletal muscle stem cells or Satellite Cells (SCs) are kept in a state of dormancy but can swiftly activate in response to external stimuli. However, the mechanisms that allow quiescent SCs to activate quickly are still unknown. In this study published in Nature Communications, Prof. Tom Cheung and his research team identified cytoplasmic polyadenylation elements-binding protein 1 (CPEB1) as a critical regulator in reprogramming the translational landscape that controls SC activation.
Tissue homeostasis and repair rely on tissue stem cells. In skeletal muscle, quiescent SCs (QSCs) are usually kept in a state of dormancy with baseline cellular metabolism. However, they can quickly activate and re-enter the cell cycle after being stimulated during injury.
Using proteomic analysis, the team led by Prof. Cheung revealed that in vivo and in vitro SCs had similar proteomic signatures after activation while QSCs preserved by in situ fixation and freshly isolated SCs (fiSCs) were more distant. They observed a considerable shift in the translational landscape for cell proliferation and metabolism reprogramming while SCs are activated.
To determine the signaling pathways in QSCs during the cell state change, solely analyzing their transcriptome is not sufficient because RNA levels may not precisely represent protein levels. From the proteomic and transcriptomic data in the study, the team found concordant upregulated or downregulated transcripts and proteins during SC activation. However, some exceptions exist in which transcripts are downregulated while proteins are still significantly expressed, suggesting that the SC quiescence-to-activation transition can be regulated post-transcriptionally to alter protein levels.
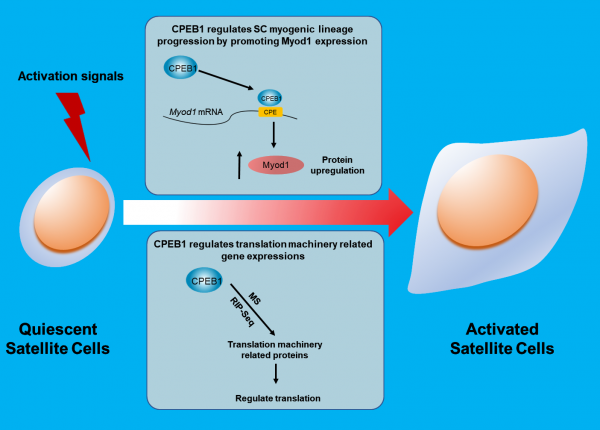
Around 20% of mammalian transcripts contain cytoplasmic polyadenylation elements (CPEs) located on the 3′ untranslated region (3′ UTR) of messenger RNA (mRNA). The CPE-binding protein 1 (CPEB1) can bind to CPE sequences and modulate translation by changing the stability of mRNAs. In this study, the Cheung lab discovered that the absence of CPEB1 in SCs reduced proliferation and caused a delay in SC activation but did not affect the fate of SC cells.
During the SC quiescence-to-activation transition, several CPEB1-associated genes were downregulated, and their protein expressions were elevated. One of these genes was the myogenic factor Myod1, required for myogenic lineage progressions. CPEB1 was shown to bind to Myod1 transcripts via CPEs and increased Myod1 protein level in a phosphorylation-dependent manner. Specifically, inhibiting CPEB1 phosphorylation reduced the percentage of Myod1+ SCs during early activation. In a muscle injury model, suppressing CPEB1 phosphorylation using pharmacological agents MK5108 dampened muscle regeneration, whereas promoting CPEB1 phosphorylation using insulin had the opposite effect.
This study revealed the transcriptomic and proteomic changes upon SC activation and identified the phosphorylation of CPEB1 as a key player in controlling protein levels during SC activation.
Journal Reference:
Zeng W, Yue L, Lam KSW, Zhang W, So WK, Tse EHY, Cheung TH. CPEB1 directs muscle stem cell activation by reprogramming the translational landscape. Nat Commun. 2022 Feb 17;13(1):947. doi: 10.1038/s41467-022-28612-1.