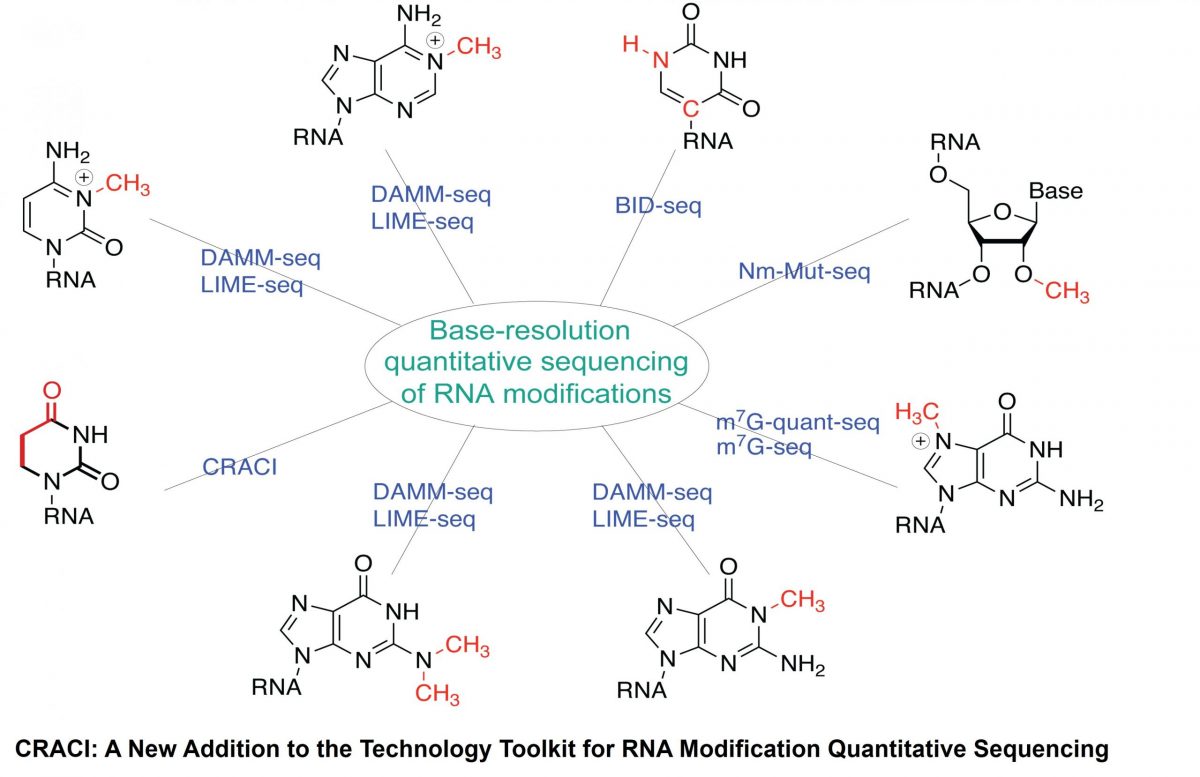
A team led by Prof. ZHANG Li‑Sheng (Division of Life Science & Department of Chemistry, HKUST) has developed Chemical Reduction Assisted Cytosine Incorporation sequencing (CRACI) — a sensitive method that quantitatively pinpoints dihydrouridine (D), the most abundant tRNA modification at the single-base resolution.
Small chemical marks on RNA—like D—can work like fine‑tuning structural switches, influencing how genes are expressed. Until now, these D marks were hard to detect. With CRACI, the team created the first detailed maps of D in cellular tRNAs from mammals and plants, including those inside mitochondria.
Key discoveries:
Because D is common and conserved across species, and related enzymes are potentially linked to human disease, CRACI gives researchers a powerful platform to explore how these RNA marks shape cell function, stress responses, and health. The team plans to extend CRACI to primary cells and tissues to build a clear and comprehensive picture of D roles in disease contexts.
Journal reference
Ju, CW., Li, H., Jiang, B. et al. Quantitative CRACI reveals transcriptome-wide distribution of RNA dihydrouridine at base resolution. Nat Commun 16, 8863 (2025). https://doi.org/10.1038/s41467-025-63918-w